All content is publicly available under the [Creative Commons Attribution License](https://creativecommons.org/licenses/by/4.0/).
:::
General questions or feedback? Contact [team@carpentries.org](mailto:team@carpentries.org).
:::warning
### Code of Conduct
We are dedicated to providing <a href="https://magic.ly/insmaifensi">INS买粉</a> a welcoming and supportive environment for all people, regardless of background or identity. By participating in this community, participants accept to abide by The Carpentries’ Code of Conduct and accept the procedures by which any Code of Conduct incidents are resolved. Any form of behaviour to exclude, intimidate, or cause discomfort is a violation of the Code of Conduct. In order to foster a positive and professional learning environment we encourage the following kinds of behaviours in all platforms and events:
- Use welcoming and inclusive language
- Be respectful of different viewpoints and experiences
- Gracefully accept constructive criticism
- Focus on what is best for the community
- Show courtesy and respect towards other community members
If you believe someone is violating the Code of Conduct, we ask that you report it to The Carpentries Code of Conduct Committee [completing this form](https://docs.google.com/forms/d/e/1FAIpQLSdi0wbplgdydl_6rkVtBIVWbb9YNOHQP_XaANDClmVNu0zs-w/viewform), who will take the appropriate action to address the situation.
See more detailed description of the Code of Conduct [here](https://docs.carpentries.org/topic_folders/policies/code-of-conduct.html).
:::
## Introduction
We are so happy to see you all here, and we look forward to the following days!
[Course page](https://korbinib.github.io/2022-12-05-UiB-swc)
[Pre-workshop survey](https://carpentries.typeform.com/to/wi32rS?slug=2022-12-05-UiB-swc)
#### This Workshop is enabled by:
[UiB Library Digital Lab](https://www.uib.no/digitallab) >> [Event calendar](https://www.uib.no/en/digitallab/calendar)
[Centre for Digital Life Norway](https://www.digitallifenorway.org/) >> [Events](https://www.digitallifenorway.org/events/)
[ELIXIR Norway](https://elixir.no/)
[Norwegian Research Infrastructure Services - NRIS](https://www.sigma2.no/nris)
#### This course is organized by:
INSTRUCTORS|HELPERS:
Illimar Rekand, illimar.rekand@uib.no, ELIXIR
Korbinian Bösl, Korbinian.Bosl@uib.no, ELIXIR & Digital Life Norway
Michael Dondrup, Elixir Norway, Computational Biology Unit
Siri Kallhovd, Siri.Kallhovd@uib.no, ELIXIR & NRIS
Gutama Ibrahim Mohammad, Gutama.Mohammad@uib.no, Computational Biology Unit
Parisa Tejari Geravand, parisa.tejari@uib.no, ELIXIR
Henrik Askjer, henrik.askjer@uib.no, Digital Lab, University Library of Bergen
David Dolan, david.dolan@uib.no, ELIXIR
Jenny Ostrop, Jenny.Ostrop@uib.no, Digital Lab, University Library of Bergen
Matúš Kalaš, Matus.Kalas@uib.no, ELIXIR
Samuel Martey Okoe-Mensah, samuel.okoe-mensah@student.uib.no
Jane Chen, hjchen619@gmail.com
Emma Josefin Ölander Aadland, emma.aadland@uib.no, Digital Lab, University Library of Bergen
Dhanya Pushpadas, Dhanya.Pushpadas@uib.no, UiB-ITA & NRIS
Please contact us if you have any questions or comments.
# Monday 25th of September
:::danger
## Schedule
### Day 1 Monday 25th of September: morning session
*Before starting: [Pre-workshop survey](https://tinyurl.com/SWC23Surv)*
[Setup](http://swcarpentry.github.io/shell-novice/setup.html) (Michael)
Download files required for the lesson
09:00 - 10:00 Installation help
10:00 The Unix Shell
**[Introducing the Shell](http://swcarpentry.github.io/shell-novice/01-intro/index.html)** (Michael)
**[Navigating Files and Directories](http://swcarpentry.github.io/shell-novice/02-filedir/index.html)** (Michael)
**[Working With Files and Directories](http://swcarpentry.github.io/shell-novice/03-create/index.html)** (Michael)
11:00 The Unix Shell (Continued)
**[Pipes and Filters](http://swcarpentry.github.io/shell-novice/04-pipefilter/index.html)** (Michael)
**[Finding Things](http://swcarpentry.github.io/shell-novice/07-find/index.html)** (Michael)
11:45 lunch
:::
### Before we start, we would like to get to know a bit more about you :)
*Please add your name to the roll call below*
Illimar Rekand
Hilde Johansen
Korbinian Boesl
Karlijn :sunny:
Louise Bjerrum
Tak Ono🌧
Haja Sherief
Andrea Campos-Candela :)
Petra Bayerova :)
T. Mutugi
Randi H Eilertsen :S
Angeliki :satellite:
Julia ;)
Leo c|\_|
Himal :)
SUNIL KUMAR PANDEY
Anna L. :P
Beatriz Diaz Pauli :)
Anna-Simone Frank :D
Liaqat Zeb
Most Champa Begum
Silje Kjølle
Kateryna
Vlad =)=
Lin
## Monday morning 25th September: The Unix Shell
## [The Unix Shell: Setup](http://swcarpentry.github.io/shell-novice/setup.html)
## [Introducing the Shell](http://swcarpentry.github.io/shell-novice/01-intro/index.html)
Q & A
* do we need to install the shell software on window now?
* on Win, the shell is included in the Git Bash - So please type along in Git Bash if you're on Win
* the *search* option is not available in --help? for windows?
* <code>man</code> in not installed by default in Git Bash on windows, but one can get the same information from *--help*. This depends on the operating system: on Mac and Linux, <code>man</code> is available by default, on Windows's Git Bash <code>man</code> is not available by default.
:::success
#### Key Points
* A shell is a program whose primary purpose is to read commands and run other programs.
* This lesson uses Bash, the default shell in many implementations of Unix.
* Programs can be run in Bash by entering commands at the command-line prompt.
* The shell’s main advantages are its high action-to-keystroke ratio, its support for automating repetitive tasks, and its capacity to access networked machines.
* The shell’s main disadvantages are its primarily textual nature and how cryptic its commands and operation can be.
:::
## [Navigating Files and Directories](http://swcarpentry.github.io/shell-novice/02-filedir/index.html)
* [Data Download for shell lessons](https://swcarpentry.github.io/shell-novice/data/shell-lesson-data.zip)
* If I am dowloading it in current Directory and extracting in the same, do I also need to move all files to desktop?
* no, you can also navigate to this directory with cd
* but remember that when the instructor says "in your Desktop" you'll have to adapt that message to your current directory...
* how to use Ctr+V In the git bash? Ctr+shift+V doesn't work
* is this in Windows? If yes, there is a soluton [here](https://stackoverflow.com/questions/2304372/how-do-you-copy-and-paste-into-git-bash).
* On Linux this might depend on your virtual terminal
* On all setups: Right click will usually either do the "paste", or show options including pasting. So it's the first option to try in each shell terminal app.
* I do not get the output I should when typing "ls -F Desktop/shell-lesson-data". I only get "shell-lesson-data/" as output. What to do?
* that is the correct outcome, good! It meas you have a directory named "shell-lesson-data" under a subdirectory named "Desktop".
* Perhaps, you were expecting something different because the instructor made a type in the name, in the first attempt, and, therefore, "ls -F" returned file not found.
* Found out I had a second folder named the same inside the shell-lesson-data folder. All good now.
* good!
* The output is identical when I write "ls" and "ls -F" in Windows. Am I doing something wrong?
* The ouput should be **almost identical** with the difference that you should see a slash "/" immediatly after the directory name, and other symbols after the names of executables, links, and other kinds of files. In practice, the only difference should be the format of the output.
* cd - doesn't work in windows. or am I doing something wrong? cd - is supposed to work as the back function? correct?
* `cd -` does bring you to the previous directory (tested on Mac/Linux/Git Bash on Win, but does NOT work in all shell terminal apps/setups).
* `cd` stands for *change directory* so depending on the argument after `cd` this command will change the directory accordingly.
* For example, `cd ..` will change the current directory to the parent directory of the current directory (more easy to see than to explain...)
* `cd` without additional argument, or `cd $HOME` or `cd ~` will move you to your home directory. But, this does NOT work on all shell terminal apps/setups.
* Is there a way to increase the font of the terminal (Git bash)?
* On Mac: press 'Command +'
* On Windows: 'Ctrl +'
* To increase the font permanently you can right click on the menu bar and open the configuration menu for the terminal of Git Bash on Windows.
:::success
### Key Points
* The file system is responsible for managing information on the disk.
* Information is stored in files, which are stored in directories (folders).
* Directories can also store other directories, which then form a directory tree.
* ```ls [path]``` prints a listing of a specific file or directory; ls on its own lists the current working directory.
* ```cd [path]``` changes the current working directory.
* ```ls [path]``` prints a listing of a specific file or directory; ls on its own lists the current working directory.
* pwd prints the user’s current working directory.
* `/` on its own is the root directory of the whole file system.
* Most commands take options (flags) that begin with a `-`.
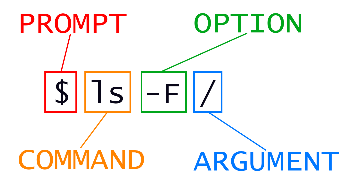
* A relative path specifies a location starting from the current location.
* An absolute path specifies a location from the root of the file system.
* Directory names in a path are separated with / on Unix, but \ on Windows.
* `..` means ‘the directory above the current one’; `.` on its own means ‘the current directory’.
### [Cheatsheet for Shell](https://cheatography.com/sjuenemann/cheat-sheets/de-nbi-workshops-linux-bash-at-a-glance/)
### [NRIS Unix-Intro documentation](https://training.pages.sigma2.no/tutorials/unix-for-hpc/)
:::
## [Pipes and Filters](http://swcarpentry.github.io/shell-novice/04-pipefilter/index.html)
**Q & A:**
* Where can I find the `|` symbol on my keyboard?
* standard QWERTY Norwegian keyboard layout: `§½` found to the left of `1!`.
* Mac QWERTY Norwegian keyboard layout: `ALT` `+` `7` - This might change if you did a `Terminal -> Settings ->Keyboard krysset av "Use Option As Meta` settings
* QWERTY English keyboard layout: `\|` found to the left of `Z`
* QWERTZ German keyboard layout: `<>|` found to the left of `Y`
* Sorting by columns in a CSV/TSV...
* https://stackoverflow.com/questions/9471101/sort-csv-file-by-multiple-columns-using-the-sort-command says:
:::warning
You need to use two options for the sort command:
`--field-separator` (or `-t`)
`--key=<start,end>` (or `-k`), to specify the sort key, i.e. which range of columns (start through end index) to sort by. Since you want to sort on 3 columns, you'll need to specify -k 3 times, for columns 2,2, 1,1, and 3,3.
To put it all together,
`sort -t ';' -k 2,2 -k 1,1 -k 3,3`
Note that sort can't handle the situation in which fields contain the separator, even if it's escaped or quoted.
:::
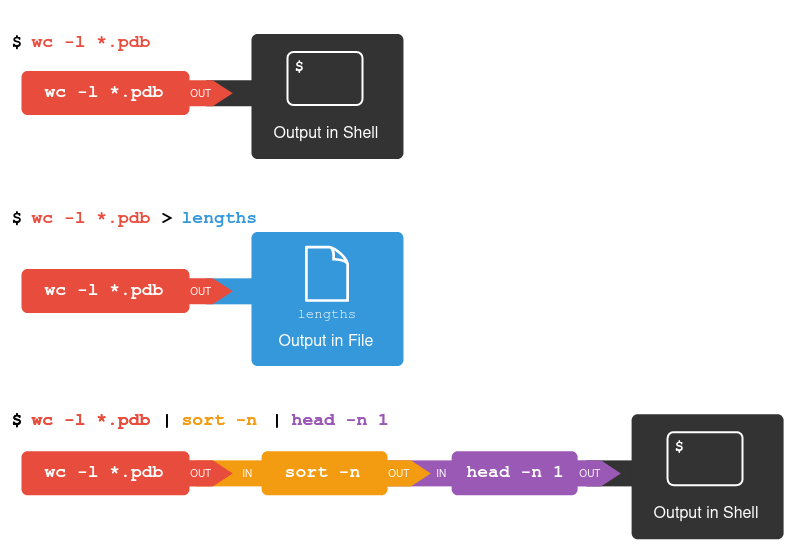
::: success
### Keypoints
* wc counts lines, words, and characters in its inputs.
* cat displays the contents of its inputs.
* sort sorts its inputs.
* head displays the first 10 lines of its input.
* tail displays the last 10 lines of its input.
* command > [file] redirects a command’s output to a file (overwriting any existing content).
* command >> [file] appends a command’s output to a file.
* [first] | [second] is a pipeline: the output of the first command is used as the input to the second.
* The best way to use the shell is to use pipes to combine simple single-purpose programs (filters).
:::
## [Loops](https://swcarpentry.github.io/shell-novice/05-loop/index.html)
Loops will be covered more extensively in the R|python lesson.
:::success
### Keypoints
* A for loop repeats commands once for every thing in a list.
* Every for loop needs a variable to refer to the thing it is currently operating on.
* Use $name to expand a variable (i.e., get its value). ${name} can also be used.
* Do not use spaces, quotes, or wildcard characters such as ‘*’ or ‘?’ in filenames, as it complicates variable expansion.
* Give files consistent names that are easy to match with wildcard patterns to make it easy to select them for looping.
* Use the up-arrow key to scroll up through previous commands to edit and repeat them.
* Use Ctrl+R to search through the previously entered commands.
* Use history to display recent commands, and ![number] to repeat a command by number.
:::
## [Shell Scripts](https://swcarpentry.github.io/shell-novice/06-script/index.html)
:::success
### Key points
* Save commands in files (usually called shell scripts) for re-use.
* bash [filename] runs the commands saved in a file.
* $@ refers to all of a shell script’s command-line arguments.
* $1, $2, etc., refer to the first command-line argument, the second command-line argument, etc.
* Place variables in quotes if the values might have spaces in them.
* Letting users decide what files to process is more flexible and more consistent with built-in Unix commands.
:::
## [Finding Things](http://swcarpentry.github.io/shell-novice/07-find/index.html)
:::success
### Key Points
* find finds files with specific properties that match patterns.
* grep selects lines in files that match patterns.
* `--help` is an option supported by many bash commands, and programs that can be run from within Bash, to display more information on how to use these commands or programs.
* `man [command]` displays the manual page for a given command.
* `$([command])` inserts a command’s output in place.
:::
## Feedback Day 1:
### Positive:
* Tålmodige veiledere, gode forklaringer underveis
* Great introduction :)
* Nice that they are many people to helpo
* Keep explaining and showing the exact steps
* Pacing, Helpersm, Support
* Course covers material from scratch: really good for beginners, documentation is really clear
* Nicely explained for non-informaticians, good helpers
### Negative:
* We went at a slow pace through the easy basic things (begin) and pretty fast through the more complicated material (end)
* Needed 1 more break in the end
* A bit more air in the room would be nice
* A bit fast at times
* Explain the commands a bit more/slower, so we can "digest" them and use them further in the seminar
* Maybe show the instructions - web page - in parallell, like next window for when we lose track of where we are
* Maybe another "wrap up" with the most important/key commands at the end
* Sometimes it was too quick away on the screen
Please get ready for tomorrow 👇🏽
# Tuesday 26th of September
## Rollcall
Korbinian - instructor for R today :)
Illimar - instructor for R aswell
Himal - R
Louise - R
Andrea Campos-Candela -- python
Angeliki - R
Julia - Python
Leo c|\_| - R
Haja Sherief (Python)
Karlijn - Python
Anna - Python
Randi - Python
Kristoffer - Python
Silje Kjølle - R
Beatriz Diaz Pauli - R
Anna-Simone -- python
Nathaniel - R
Mutugi -R
Lin -R
Kateryna R
Liaqat Zeb R
Petra Bayerova R
Matúš Kalaš - Python (helper)
Christiane Eichner - Python
Most Champa Begum - Python
Jenny Ostrop - R (helper)
Sunil Kumar Pandey (Does´nt have the git)
)
Total: 23
# R
:::danger
## Schedule
### Day 2 Tuesday 26th: morning session
[Setup](https://swcarpentry.github.io/r-novice-gapminder/setup.html) (Korbinian)
R for Reproducible Scientific Analysis
**[Introduction to R and RStudio](https://swcarpentry.github.io/r-novice-gapminder/01-rstudio-intro.html)** (Illimar)
**[Project Management With RStudio](https://swcarpentry.github.io/r-novice-gapminder/02-project-intro.html)** (Korbinian)
**[Seeking Help](https://swcarpentry.github.io/r-novice-gapminder/03-seeking-help.html)** (Illimar)
*10:30 Break*
10:45 R for Reproducible Scientific Analysis (Continued)
**[Data Structures](https://swcarpentry.github.io/r-novice-gapminder/04-data-structures-part1.html)** (Korbinian)
12:00 Lunch
:::
Please make sure you have R and RStudio installed for later. [Please see the setup information🙂](https://korbinib.github.io/2022-12-05-UiB-swc/#r)
If possible, please also install some R packages for later. On Mac/Linux in your bash type (after installing R):
```R -e 'install.packages("tidyverse",repos = "https://cran.uib.no/")'```
On Windows you can install them in RStudio, for example by creating a script _prepare_installation.r_:
```
install.packages("tidyverse")
library(tidyverse)
```
*****
## R installation
[Setup instructions](https://swcarpentry.github.io/r-novice-gapminder.html)
## [Introduction to R and RStudio](https://swcarpentry.github.io/r-novice-gapminder/01-rstudio-intro.html)
**[Cheatsheet R](https://github.com/rstudio/cheatsheets/raw/main/base-r.pdf)**
Different people use different conventions for long variable names, these include
* periods.between.words
* underscores_between_words
* camelCaseToSeparateWords
What you use is up to you, but be **consistent**.
[Tidyverse Style Guide](https://style.tidyverse.org/syntax.html)
[Google's R Style Guide](https://google.github.io/styleguide/Rguide.html)
:::warning
### Challenge 1
Which of the following are valid R variable names? add 😀 | 😞 behind each
min_height
max.height
_age
.mass
MaxLength
min-length
2widths
celsius2kelvin
:::
**Q & A:**
* what is the difference between = and <- assignments to variables?
* in function arguments you can only use `=`; other than that there are no major differences.
* I cannot clean the console on my MAC, tried press option command L as well..
* Try Control small l
* Worked! thanks
* you can also do it with the mouse 
* I am installing packages, and it is taking forever.. Can't do much now.
* to assign y to 2*x really, would one have to make a function? or maybe use symbolic?
* Using x <- 2*x will assign a static variable (previous value of x times 2), for a different behaviour functions will help you if you want to do this more often
* What is the difference coding in the consol vs coding in the file window?
* none, but you can save the file to rerun it (as a script) later :) - Your console is not persistent
* from where to open descriptive window?
* assuming this is about the file view: click the file -> new file -> R script
* don't you usually install packages in the script instead of console?
* no, as you only need to install them once. You will need to make them available with library(packagename) which is often done in the script. We will get to this.
* WARNING: Rtools is required to build R packages but is not currently installed. Please download and install the appropriate version of Rtools before proceeding: https://cran.rstudio.com/bin/windows/Rtools/ Warning in install.packages : 'lib = "packages"' is not writable
* I guess you should install Rtools before. Please follow the instructions here: https://cran.r-project.org/bin/windows/Rtools/
* For gapminder installation, it says - Rtools is required
* on Windows try installing https://cran.r-project.org/bin/windows/Rtools/
* How can you be sure that the package is downloaded successfully?
* You can try to load it, for example library(tidyverse)
* what should happen in the console then? sorry, I mean what happens if you load with library (tidyverse)? If it comes up as an option with purple
* Could you specify?
* Depends on the package. If you do not get an error message it's usually good. Some packages (as tidyverse) will show you extra information.
* Ok, that make sense:)
:::success
### Key Points
* Use RStudio to write and run R programs.
* R has the usual arithmetic operators and mathematical functions.
* Use <- to assign values to variables.
* Use ls() to list the variables in a program.
* Use rm() to delete objects in a program.
* Use install.packages() to install packages (libraries).
:::
We will work with the gapminder data set with is the basis for [this inspiring talk from Hans Rosling 20min](https://www.youtube.com/watch?v=hVimVzgtD6w).
[4min short version](https://www.youtube.com/watch?v=Z8t4k0Q8e8Y)
## [Project Management With RStudio](https://swcarpentry.github.io/r-novice-gapminder/02-project-intro/index.html)
:::warning
### Challenge 1+2: Creating a self-contained project
We’re going to create a new project in RStudio:
1. Click the “File” menu button, then “New Project”.
2. Click “New Directory”.
3. Click “New Project”.
4. Type in the name of the directory to store your project, e.g. “my_project”.
5. If available, select the checkbox for “Create a git repository.”
6. Click the “Create Project” button.
### ...and open it again
1. Exit RStudio.
2. Navigate to the directory where you created a project in Challenge 1.
3. Double click on the `.Rproj` file in that directory.
:::
Link for Gapminder data: https://swcarpentry.github.io/r-novice-gapminder/data/gapminder_data.csv
:::success
### Key Points
* Use RStudio to create and manage projects with consistent layout.
* Treat raw data as read-only.
* Treat generated output as disposable.
* Separate function definition and application.
:::
## [Seeking Help](https://swcarpentry.github.io/r-novice-gapminder/03-seeking-help/index.html)
### R SessionInfo
R version 4.1.2 (2021-11-01)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur 11.5.2
R version 4.2.3 (2023-03-15 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
R version 4.3.1 (2023-06-16 ucrt) -- "Beagle Scouts"
Platform: x86_64-w64-mingw32/x64 (64-bit)
:::success
### Key Points
* Use `help()` to get online help in R.
* Use `?function-name` to read the documentation
* Use `??function-name` if you are not sure about spelling
* Read vignettes to learn more about usage
* Use `sessionInfo()` to get information about your R and package versions
:::
ELIXIR-NO has a helpdesk that can help you with your scripting for Life Science qqqhttps://elixir.no/helpdesk
## [Data Structures](https://swcarpentry.github.io/r-novice-gapminder/04-data-structures-part1/index.html)
```
cats <- data.frame(coat = c("calico", "black", "tabby"),
weight = c(2.1, 5.0, 3.2),
likes_strings = c(1, 0, 1))
```
There are 5 main types in R: **double, integer, complex, logical, and character.**
:::warning
Order of datatypes in R: **logical -> integer -> numeric -> complex -> character**
:::
:::success
### Key points
* Use `read.csv` to read tabular data in R.
* The basic data types in R are double, integer, complex, logical, and character.
* Use factors to represent categories in R.
:::
:::danger
# Schedule
## Day 2 Tuesday 26th: afternoon session
**[Exploring Data Frames](https://swcarpentry.github.io/r-novice-gapminder/05-data-structures-part2.html)** (Illimar)
**[Subsetting Data](https://swcarpentry.github.io/r-novice-gapminder/06-data-subsetting.html)** (Korbinian)
**[Control Flow](https://swcarpentry.github.io/r-novice-gapminder/07-control-flow.html)** (Korbinian)
**[Creating Publication-Quality Graphics with ggplot2](https://swcarpentry.github.io/r-novice-gapminder/08-plot-ggplot2.html)** (Illimar)
**[Data frame Manipulation with dplyr](https://swcarpentry.github.io/r-novice-gapminder/13-dplyr.html)** (Korbinian)
**Tidy data, wide and long tables** (Korbinian)
**[Writing Good Software](https://swcarpentry.github.io/r-novice-gapminder/16-wrap-up.html)** (Illimar)
*16:00 END*
:::
## [Exploring Data Frames](https://swcarpentry.github.io/r-novice-gapminder/05-data-structures-part2.html)
:::warning
### Challenge
* Create a vector with numbers 1-26, then multiply by 2
my_vector <- 1:26
my_multiplied_vector <- my_fantastic_vector * 2
* Let's imagine that 1 cat year is equivalent to 7 human years
1. Create a vector called human_age by multiplying cats$age by 7
2. Convert human_age to a factor
3. Convert human_age back to a numeric vector using the as.numeric(function). Now divide it by 7 to get the original ages back. Explain what happened.
:::
:::warning
You can create a new data frame right from within R with the following syntax:
R
df <- data.frame(id = c("a", "b", "c"),
x = 1:3,
y = c(TRUE, TRUE, FALSE))
Make a data frame that holds the following information for yourself:
first name
last name
lucky number
:::
Starting with the gapminder dataset!
```gapminder <- read.csv("data/gapmnder_data.csv")```
Use ```str(gapminder)```, ```summary(gapminder)``` to get an overview about the data and try different functions such as ```dim()```, ```colnames()```, ```typeof()``` to explore the dataset.
### Key Points
* Use `cbind()` to add a new column to a data frame.
* Use `rbind()` to add a new row to a data frame.
* Remove rows from a data frame.
* Use `na.omit()` to remove rows from a data frame with NA values.
* Use `levels()` and `as.character()` to explore and manipulate factors.
* Use `str()`, `summary()`, `nrow()`, `ncol()`, `dim()`, `colnames()`, `rownames()`, `head()`, and `typeof()` to understand the structure of a data frame.
* Read in a csv file using `read.csv()`.
* Understand what `length()` of a data frame represents.
## [Subsetting Data](https://swcarpentry.github.io/r-novice-gapminder/06-data-subsetting.html)
:::info
### Vector numbering in R starts at 1
In many programming languages (C and Python, for example), the first element of a vector has an index of 0. In R, the first element is 1.
:::
:::info
### Tip: Non-unique names
You should be aware that it is possible for multiple elements in a vector to have the same name. (For a data frame, columns can have the same name — although R tries to avoid this — but row names must be unique.) Consider these examples:
```
x <- 1:3
x
```
**Output**
```
[1] 1 2 3
```
```
names(x) <- c('a', 'a', 'a')
x
```
**Output**
```
a a a
1 2 3
```
```
x['a'] # only returns first value
```
**Output**
```
a
1
```
```
x[names(x) == 'a'] # returns all three values
```
**Output**
```
a a a
1 2 3
```
:::
:::warning
### Challenge 3
Selecting elements of a vector that match any of a list of components is a very common data analysis task. For example, the gapminder data set contains country and continent variables, but no information between these two scales. Suppose we want to pull out information from southeast Asia: how do we set up an operation to produce a logical vector that is TRUE for all of the countries in southeast Asia and FALSE otherwise?
Suppose you have these data:
```R
seAsia <- c("Myanmar","Thailand","Cambodia","Vietnam","Laos")
## read in the gapminder data that we downloaded in episode 2
gapminder <- read.csv("data/gapminder_data.csv", header=TRUE)
## extract the `country` column from a data frame (we'll see this later);
## convert from a factor to a character;
## and get just the non-repeated elements
countries <- unique(as.character(gapminder$country))
```
There’s a wrong way (using only ==), which will give you a warning; a clunky way (using the logical operators == and |); and an elegant way (using %in%). See whether you can come up with all three and explain how they (don’t) work.
:::
The best way to do this problem is ```countries %in% seAsia```, which is both correct and easy to type (and read).
*We will look further into subsetting tomorrow. If you have not enough yet, take a look at challenge 7 and 8.*
:::success
### Key Points
* Indexing in R starts at 1, not 0.
* Access individual values by location using `[]`.
* Access slices of data using `[low:high]`.
* Access arbitrary sets of data using `[c(...)]`.
* Use logical operations and logical vectors to access subsets of data.
:::
## [Control Flow](https://swcarpentry.github.io/r-novice-gapminder/07-control-flow.html)
We want to compare life expectancy between continents in the gapminder dataset. To do this, we need a few tools.
If...else conditions follow the pattern:
```
# if
if (condition is true) {
perform action
}
# if ... else
if (condition is true) {
perform action
}
else if (other condition is true){
perform alternative action (optional)
}
else { # that is, if the condition(s) is/are false
perform alternative action
}
```
Sometimes, it is useful to use ```any()``` and ```all() ```in the conditions.
For loops follow the pattern:
```
# for loop
for (iterator in set of values) {
do a thing
}
# example
for (i in 1:10) {
print(i)
}
```
**Functions**
From here, it is only a small step to wrapping your script in a function with defined inputs and outputs that you can reuse. Functions follow the pattern:
```
my_function <- function(parameters) {
# perform action (e.g for loop, if...else condition)
# return value
}
```
You can read more about writing functions in R in the [Functions explained](https://swcarpentry.github.io/r-novice-gapminder/10-functions.html) lesson. We recommend to solve Challenge 1 and Challenge 2 as homework. You can find a model solution in the [Functions explained](https://swcarpentry.github.io/r-novice-gapminder/10-functions.html) lesson.
:::success
### Key Points
* Use `if` and `else` to make choices.
* Use `for` to repeat operations.
* Use `function` to define a new and reusable functions in R.
* Use parameters to pass values into functions.
:::
:::warning
### Challenge 1 (recommended homework)
Write a function called ```kelvin_to_celsius()``` that takes a temperature in Kelvin and returns that temperature in Celsius.
Hint: To convert from Kelvin to Celsius you subtract 273.15
:::
:::warning
### Challenge 2 (recommended homework)
Define the function to convert directly from Fahrenheit to Celsius, by reusing the two functions above (or using your own functions if you prefer).
:::
## [Creating Publication-Quality Graphics with ggplot2](https://swcarpentry.github.io/r-novice-gapminder/08-plot-ggplot2/index.html)
**Introducting Tidyverse**
You might have noticed that deviated from the lesson material by asking you to install the ```tidyverse``` package instead of installing the packages ```ggplot2```, ```dplyr``` and ```tidyr```. These, and other packages, are part of the```tidyverse``` package. ```Tidyverse``` is "an opinionated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures." You can read more about ```tidyverse``` [here](https://www.tidyverse.org/).
Yesterday and this morning, we have been following the Base-R syntax. Knowing about the principles and quirks of R is very useful when you start working with your own data and doing more complex things. For the rest of today, we will take a look at the```tidyverse``` grammar.
**Getting started with ggplot**
Gg stands for "grammar of graphics".
NB! The example plots are not exactly publication grade. A few **tricks & useful links to make plots more appealing**:
[**Cheatsheet ggplot2**](https://raw.githubusercontent.com/rstudio/cheatsheets/main/data-visualization.pdf)
[R graph gallery & code snippets](https://www.r-graph-gallery.com/index.html)
[ggplot2 themes](https://ggplot2.tidyverse.org/reference/ggtheme.html)
[Color palettes](https://colorbrewer2.org/)
[Scientific journal and sci-fi themes](https://cran.r-project.org/web/packages/ggsci/vignettes/ggsci.html)
[Arranging plots with cowplot](https://cran.r-project.org/web/packages/cowplot/vignettes/introduction.html)
[ggplot2 book](https://ggplot2-book.org/)
(available at the UiB library: https://bibsys-almaprimo.hosted.exlibrisgroup.com/permalink/f/1cruloh/BIBSYS_ILS71500376670002201)
[Fundamentals of Data Visualization book](https://clauswilke.com/dataviz/)
(available at the UiB library: https://bibsys-almaprimo.hosted.exlibrisgroup.com/permalink/f/8hnp7t/BIBSYS_ILS71576188470002201)
[Examples of Different Figure Types with Code] (https://www.data-to-viz.com/)
:::warning
### Challenge 5
Generate boxplots to compare life expectancy between the different continents during the available years.
Advanced:
Rename y axis as Life Expectancy.
Remove x axis labels.
:::
:::success
### Key Points
* Use ```ggplot2``` to create plots.
* Save you plots with ````ggsave```
* Think about graphics in layers: aesthetics, geometry, statistics, scale transformation, and grouping.
* Make use of cheatsheets, documentation, vignettes, and other resources.
:::
## [Data frame Manipulation with dplyr](https://swcarpentry.github.io/r-novice-gapminder/13-dplyr/index.html)
select()
filter()
group_by()
summarize()
mutate()
:::warning
%>% (pipe) shortcut: Ctrl +Shift +M
:::
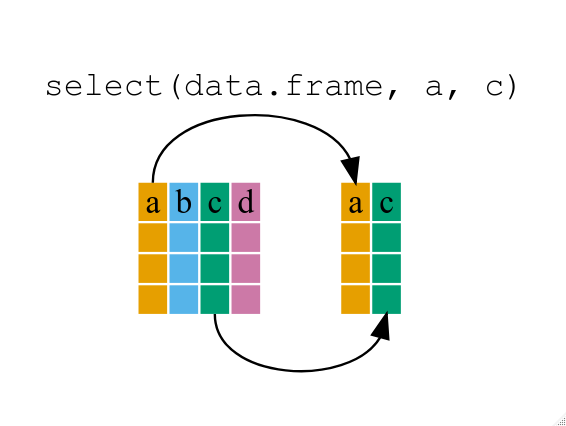
[**dplyr cheatsheet**](https://posit.co/wp-content/uploads/2022/10/data-transformation-1.pdf)
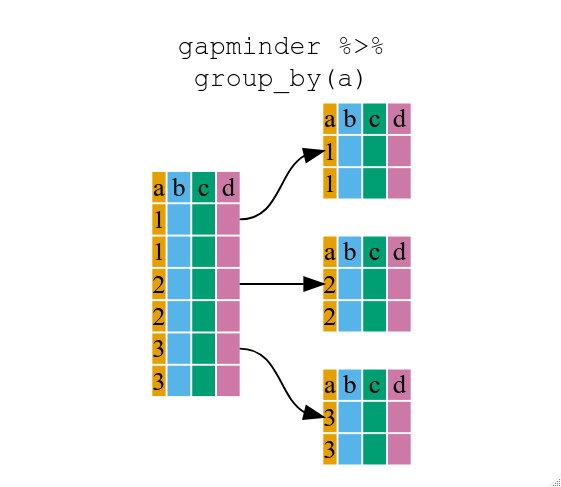
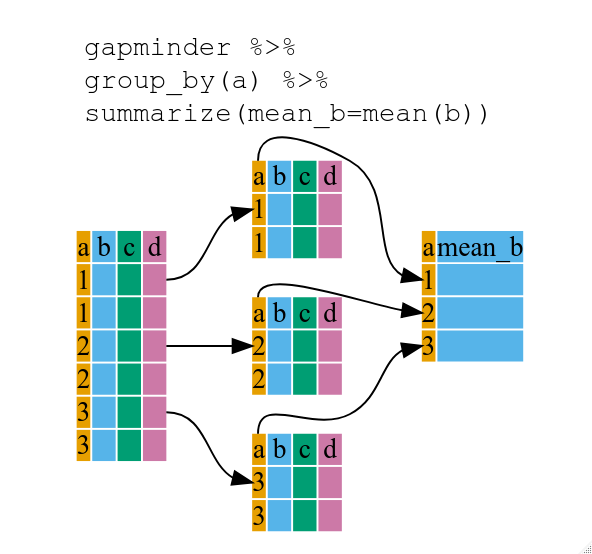
:::success
### Key Points
* Use the dplyr package to manipulate data frames.
* Use select() to choose variables from a data frame.
* Use filter() to choose data based on values.
* Use group_by() and summarize() to work with subsets of data.
* Use mutate() to create new variables.
:::
**Further resources**
* [R for Data Science](http://r4ds.had.co.nz/)
* [Data Wrangling Cheat sheet](https://www.rstudio.com/wp-content/uploads/2015/02/data-wrangling-cheatsheet.pdf)
* [Introduction to dplyr](https://dplyr.tidyverse.org/)
* [Data wrangling with R and RStudio](https://www.rstudio.com/resources/webinars/data-wrangling-with-r-and-rstudio/)
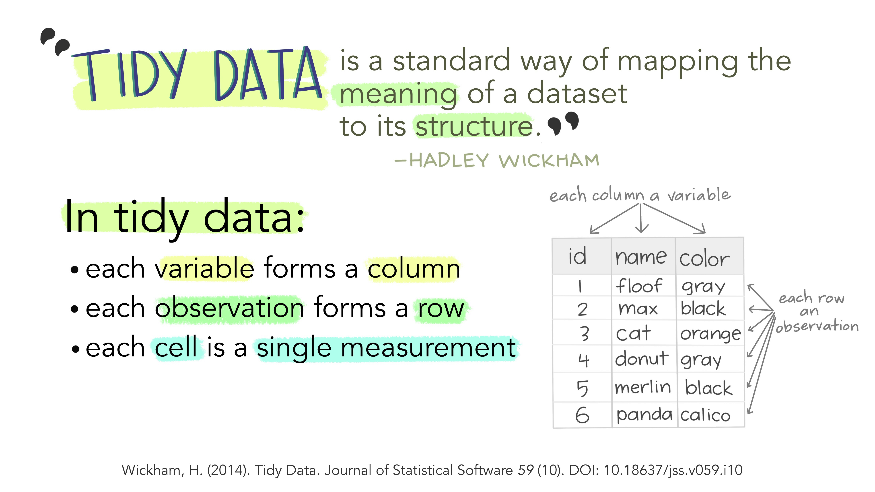
## Tidy data, wide and long tables
Do you remember the manifesto that we looked at when checking ```vignette(package="tidyverse")```? One central concept in ```tidyverse``` is the reusability of data structures.
(illustration by [Allison Horst](https://github.com/allisonhorst/stats-illustrations))
Ggplot2 plots work best with data in the *long* format, i.e., a column for every variable, and a row for every observation. But what if you have several variables? The solution is to "wrangle" the data into a longtable.
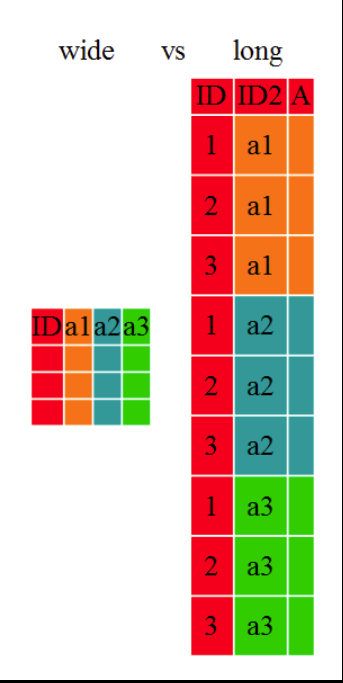
The tidyr package allows to conviently reformat data from *wide* to *long* format and back. Transformation of *wide* tables to *long* tables using the ```tidyr::pivot_longer()``` function and back using the ```tidyr::pivot_wider()``` function is explained in the [Data Frame Manipulation with tidyr](https://swcarpentry.github.io/r-novice-gapminder/14-tidyr/index.html) lesson (these functions were called ```tidyr::gather()``` & ```tidyr::spread()```earlier, which you may still find in older cheatsheets).
[**Cheatsheet data wrangling with dplyr & tidyr**](https://www.rstudio.com/wp-content/uploads/2015/02/data-wrangling-cheatsheet.pdf)
[**Cheatsheet tidyverse for beginners**](https://images.datacamp.com/image/upload/v1676302697/Marketing/Blog/Tidyverse_Cheat_Sheet.pdf)
## [Writing Good Software](https://swcarpentry.github.io/r-novice-gapminder/16-wrap-up/index.html)
:::success
### Key Points
* Keep your project folder structured, organized and tidy.
* Document what and why, not how.
* Break programs into short single-purpose functions.
* Write re-runnable tests.
* Don’t repeat yourself.
* Be consistent in naming, indentation, and other aspects of style. (have a look at the style resources shared yesterday!)
:::
:::info
## Skipped lessons R
**[9. Vectorization](https://swcarpentry.github.io/r-novice-gapminder/09-vectorization.html)**
We mentioned that vectorization is a powerful behaviour if calculating with R. This lesson goes more into depth.
**[10. Functions Explained](https://swcarpentry.github.io/r-novice-gapminder/10-functions.html)**
We have briefly talked about functions in the lesson about if...else statements and for loops.
**[11. Writing Data (in more depths)](https://swcarpentry.github.io/r-novice-gapminder/11-writing-data.html)**
We have read in and written data in different lessons, this one is a summary.
**[12. Splitting and Combining Data Frames with plyr](https://swcarpentry.github.io/r-novice-gapminder/12-plyr.html)**
More tidyverse grammar.
**[14. Data Frame Manipulation with tidyr](https://swcarpentry.github.io/r-novice-gapminder/14-tidyr.html)**
More tidyverse grammar. We have briefly talked about tidyr features in the context of tidy data and wide and long tables.
**[15. Producing Reports With knitr](https://swcarpentry.github.io/r-novice-gapminder/15-knitr-markdown.html)**
Writing reports with embedded R code and plots.
A popular alternative for embedded code and plots are [Jupyter Notebooks](https://jupyter.org/).
:::
**Further resources:**
Bioscientific packages: https://www.bioconductor.org/
https://cran.r-project.org/web/packages/available_packages_by_name.html
Next level: https://coderefinery.org/
[Coderefinery YouTube Channel](https://www.youtube.com/@coderefinery3414)
For further Training notification subscribe to [hpcnews@uib](https://mailman.uib.no/listinfo/hpcnews)
**Community in Bergen:**
* [RLadies Bergen](https://www.meetup.com/rladies-bergen/?_cookie-check=EEFbvku63iQtPzVC)
* [BioCeed R coding club](https://coderclub.w.uib.no/)
# Python
[Course page](https://swcarpentry.github.io/python-novice-gapminder/instructor/index.html)
[Download the data](https://swcarpentry.github.io/python-novice-gapminder/files/python-novice-gapminder-data.zip)
:::info
Jupyter allows you to run code cells in any order you want, but this can cause unexpected results if you want to run the code again later. Therefore it is a good idea to write the code in the order you intend to run it.
:::
## Feedback Day 2:
### General
* God hjelp å få, lett å spørre om ting, lavterskel atmosfære
* Thank you for preparing this session! It was packed with lots of new infromation for me
### R
#### Positive:
* comperatively easy to follow up than day 1
* The second part (after lunch) was efficient and interesting - using "real data"
#### Negative:
* In the R session: Most of the day was used to show basic R (subset, substract, for loop etc) to later explain that the same can be done in a more intuitive way with tidyverse. Maybe teaching only tidyverse would be more time-efficient
* Ikke nok pauser, litt ustrukturert, lite forklaring av funksjoner som ble brukt underveis (eks c() ), hjelp kom litt sent noen ganger
* 1) Need more breaks in between, 2) more time and more detail explaining the practical part with the real data, 3) More time to understand the codes during class
* First part (before lunch) was a bit slow and repetitive. Maybe just use the gapminder dataset from the beginning. Also, maybe show fewer examples
* I think maybe it should be a 5-day program OR more time for the "after-lunch" topics? maybe beginner/novice and 'intermediate' tracks that go in parallell?
### Python
#### Positive
* Very good explanation especially in the first part, quick help
* Good support from the helpers
* Very good tempo, the instructor handled the rythm very well <3
#### Negative
* a bit rushed in the end
* Room downstairs had a bad air quality and some noise from the students
* Due to time limit, bit rushed in the end
Please get ready for tomorrow 👇🏽
https://swcarpentry.github.io/git-novice
# Wednesday 27th of September
:::success
❗ After you arrive to the room, please check the basic setup at https://swcarpentry.github.io/git-novice
:::
## Rollcall & installation test
Please write your name, your scientific domain, and git version. You'll get it using the following command in your "terminal", then copy & paste from there to here:
`git --version`
__Name | Scientific domain | Git version__
Matúš Kalaš (teaching Git this morning😊) | Informatics for science, ontologies | git version 2.42.0.windows.2
Himal | Medical Biology | git version 2.33.0 (Mac) :
Illimar (does not have git on his iPad :( ) - organiser
Beatriz | Biology | git version 2.33.0 (mac M1)
Karlijn | Earth Sciences | git version 2.33.0 (Mac M1)
Hilde | Terminology/linguistics | git version 2.42.0.2 (Windows)
Lin | Medicine | git version 2.42.0.2
Nathaniel | Biology | git version 2.42.0 (Mac Intel)
Louise | cognitive neuroscience and cog. psychology | git version 2.42.0.windows.2
Silje Kjølle | Molecular biology | git version 2.42.0.windows.2
Haja Sherief, Engineering Computing, git 2.42.0.windows.2
Christiane Eichner | Biology | git version 2.30.0.windows.2
Andrea Campos-Candela | Biology | git version 2.42.0 (windows)
Most Champa Begum | Clinical Medicine | git version 2.39.1.windows.1
Jenny Ostrop (helper; teaching this afternoon: open science) | Library/Data Management, Molecular Biology | git version 2.38.1.windows.1
Siri Kallhovd (teaching Git and GitHub today) | Informatics, development, | git version 2.39.3 (Apple Git-145)
Randi H Eilertsen | git version 2.42.0.windows.2
Mutugi |git version 2.42 Windows.2
Petra | political science | git version 2.42.0 | windows
Korbinian Bösl (helper) | Data management, Molecular Biology/Bioinformatics | git 2.42.0 | linux
Parisa Tejari (helper) | Computational Biology Unit | git version 2.17.1| Ubuntu
Tak Ono | System Dynamics | git version 2.42.0.windows.2
Dhanya Pushpadas (helper) | IT Department | git version 2.38.1 | mac
David Dolan (helper)
# Git
The programme of today is to follow the lesson of https://swcarpentry.github.io/git-novice
The blue headings are links to the corresponding sections of the Git lesson.
:::info
### Schedule
Matus:
- __[1. Version control](https://swcarpentry.github.io/git-novice/01-basics.html)__
- __[2. Setting up Git](https://swcarpentry.github.io/git-novice/02-setup.html)__
Siri:
- __[3. Creating a Git repository](https://swcarpentry.github.io/git-novice/03-create.html)__
- __[4. Tracking changes](https://swcarpentry.github.io/git-novice/04-changes.html)__
Dhanya: Next training courses by NRIS __https://www.sigma2.no/training__
11:45 LUNCH
___
__12:30 RESTART__
Matus:
- __[5. Exploring history](https://swcarpentry.github.io/git-novice/05-history.html)__
- __[6. Ignoring some files](https://swcarpentry.github.io/git-novice/06-ignore.html)__
Siri:
- __[7. Using GitHub](https://swcarpentry.github.io/git-novice/07-github.html)__
- __[8. Collaborating](https://swcarpentry.github.io/git-novice/08-collab.html)__
- __[9. Conflicts](https://swcarpentry.github.io/git-novice/09-conflict.html)__
Jenny (short chapters):
- __[10. Open science](https://swcarpentry.github.io/git-novice/10-open.html)__
- __[11. Licensing](https://swcarpentry.github.io/git-novice/11-licensing.html)__
- __[12. Citation](https://swcarpentry.github.io/git-novice/12-citation.html)__
- __[13. Hosting](https://swcarpentry.github.io/git-novice/13-hosting.html)__
- __[Bonus: Using Git in RStudio](https://swcarpentry.github.io/git-novice/14-supplemental-rstudio.html)__
:::
## [1. Version control](https://swcarpentry.github.io/git-novice/01-basics.html)
:::success
### Extra exercise, if you have time
Think about a Google Document that you worked on together with many other people. Try to find it in your GoogleDocs history.
When you found it, explore the changes and previous versions via `File > Version history > See version history`
_Note: The above is only possible if you have a __"write"__ permission to the document._
Can you find out who wrote which part? And when?
☠ Do not click `Restore this version`❗
Instead, you can `Make a copy`.
__Spoiler of the day:__ All of the above will work much better (and __safer__) with Git 🙌🏽
:::
## [2. Setting up Git](https://swcarpentry.github.io/git-novice/02-setup.html)
:::danger
### Experts' trick
Run the following to see all of your Git configuration, and where it is set up:
`git config --list --show-origin`
This is the first aid when __troubleshooting!__ 😉
Other useful troubleshooting commands:
`git --version`
`pwd`
`where git`
`which git`
:::
:::info
### Cheatsheet for Git
`git help`
Running __git help__ will show you the main Git commands, with a very short description.
There are other Git cheatsheets online, more comprehensive but then also less concise...
:::
## [3. Creating a Git repository](https://swcarpentry.github.io/git-novice/03-create.html)
`git init`
`git status`
## [4. Tracking changes](https://swcarpentry.github.io/git-novice/04-changes.html)
`git add <file>`
`git commit`
`git log`
`git diff`
## [5. Exploring history](https://swcarpentry.github.io/git-novice/05-history.html)
`git show` shows the details of the last commit, or the commit we're looking at right now(!)
`git checkout <commit/branch> <file>` is an older command for discarding changes. __The modern command is__ `git restore` 💡
`git checkout <commit/branch>` is an older command for switching to another commit or branch. __The modern command is__ 'git switch' 💡
## [6. Ignoring some files](https://swcarpentry.github.io/git-novice/06-ignore.html)
`nano .gitignore`
## [7. Using GitHub](https://swcarpentry.github.io/git-novice/07-github.html)
:::danger
### [Set up an SSH key](https://swcarpentry.github.io/git-novice/07-github.html#create-an-ssh-key-pair)
`ls -la ~/.ssh` to see if you have an SSH key already
`ssh-keygen -t ed25519 -C "some blah blah"` to generate an SSH key. You can keep the defalut name of othe file (just press Enter), but afterwards create a password to access the key, and repeat the password.
__Copy the PUBLIC key, ☠ NOT THE PRIVATE ❗:__ `cat ~/.ssh/id_ed25519.pub`
:::
## [8. Collaborating](https://swcarpentry.github.io/git-novice/08-collab.html)
**Q&A**
*
*
*
## [9. Conflicts](https://swcarpentry.github.io/git-novice/09-conflict.html)
**Q&A**
*
*
*
## Open Science benefits
* Reproducibility enhances trust in science
* Increased visibility & wider audience (inside & outside academia)
* More citations (publications & data sets)
* Collaborations, others can build on your work and credit you
* Early feedback (on pre-registration, preprints, code etc.)
:::info
- 10. **[Lesson Open science](https://swcarpentry.github.io/git-novice/10-open/index.html)**
:::
### Reproducible & citable code

Modified from Katz et al. 2021 (CC BY); Five recommendations for FAIR software: https://fair-software.eu/
* Public repository with version control
* Software license: choosealicense.com (Include LICENSE.md file in repository)
* Archive code & enable citation (Include [CITATION.cff](https://docs.github.com/en/repositories/managing-your-repositorys-settings-and-features/customizing-your-repository/about-citation-files) file in repository)
* Recording of dependencies and environments Software quality measures
### Further resources
:::info
- 11. **[Lesson Licensing](https://swcarpentry.github.io/git-novice/11-licensing/index.html)**
- 12. **[Lesson Citation](https://swcarpentry.github.io/git-novice/12-citation/index.html)**
:::
[Sharing research data and software, doi 10.5281/zenodo.8086413](https://doi.org/10.5281/zenodo.8086413)
[UiB Library Open Science webinars (calendar)](https://www.uib.no/en/ub/calendar)
Life Science Data Management courses by [ELIXIR Norway](https://elixir.no/)
## Skipped lessons Git:
:::info
- 13. **[Hosting](https://swcarpentry.github.io/git-novice/13-hosting/index.html)**
- 14. **[Using Git from RStudio](https://swcarpentry.github.io/git-novice/14-supplemental-rstudio/index.html)**
:::
I found this resource to be very helpful in getting github set up with R and RStudio: https://happygitwithr.com/index.html
**Q & A:**
*
*
## Summary (cheat sheet)
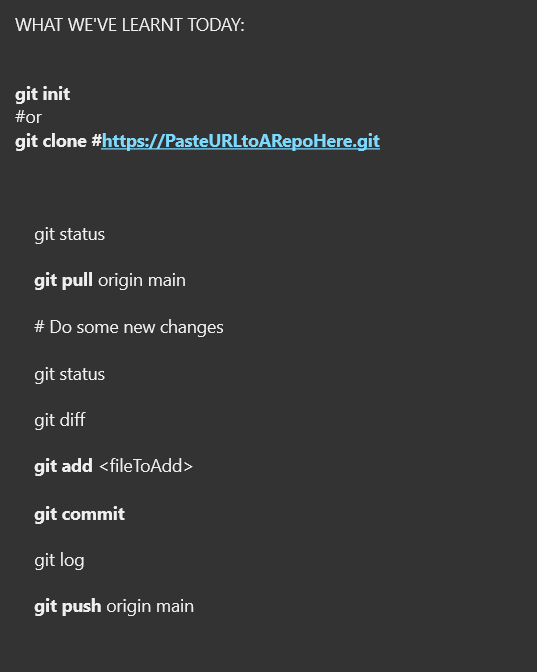
****
****
## Wrap-up
### Find out about future training events
* UiB Library Digital Lab [calendar](https://www.uib.no/en/digitallab/calendar) and mailing list (digital.lab.ub@uib.no) >> to be added, send email to Digital Lab coordinator Emma Josefin Ölander Aadland <Emma.Aadland@uib.no>
* [ELIXIR-NO](https://elixir.no/) and [Centre for Digital Life Norway](https://www.digitallifenorway.org/) provide various courses on research data management and skills for (digital) life sciences
* check out also our [research school](https://www.digitallifenorway.org/research-school/membership/index.html)
* UiB IT & NRIS Training Announcements [Subscribe to hpcnews](https://mailman.uib.no/listinfo/hpcnews)
:::success
## Next level: [**CodeRefinery**](https://coderefinery.org/)
We strongly recommend checking out [**CodeRefinery**](https://coderefinery.org/), to get more training on:
- **Git** (including the repetition of the very basics, plus collaboration with **branches** and pull requests)
- **Open science**, **licensing**, **reproducible research**
- **Documentation**, **testing**, **writing better code**, *etc.* (independent of a concrete programming language)
The educational [materials from **CodeRefinery**](https://coderefinery.github.io/2023-09-19-workshop/) are also good for **self-learning** ([__with video__](https://coderefinery.github.io/2023-09-19-workshop/) if you prefer, but also very nice to follow without watching the video recordings). **Or join an online workshop!** 🙌🏽
(Now that you know the basics, you should consider whether you join a CodeRefinery workshop as a [__helper__ a.k.a. __team leader__](https://coderefinery.github.io/2023-09-19-workshop/join/) 😉)
:::
## Feeback for Wednesday and/or the whole course
:::danger
### **Please fill out the [Post-workshop survey](https://carpentries.typeform.com/to/UgVdRQ?slug=2023-09-25-UiB-swc-!)**
:::
Please let us know what we should continue with/improve:
*
*
*
*
*
*